Urgent Info
- There is no Urgent Info now.
GSIC
| Addr. | 2-12-1 O-okayama, Meguroku, Tokyo 152-8550 JAPAN |
| Contact this mail address. |
You are here
Multiphysics Biofluidics Simulation
Massimo Bernaschi, IAC, National Resource Counsil, Italy
Satoshi Matsuoka, GSIC, Tokyo Institute of Technology
Background and Overview
Gaining a deeper insight of blood circulation is a necessary step to improve our understanding of cardiovascular diseases. However, a detailed, realistic simulation of hemodynamics represents challenging issues both in terms of physical modeling and high performance computing technology. The model must handle both the motion of fluid within the complex geometry of the vasculature, which may be unsteadily changed by heartbeat, and the dynamics of red blood cells (RBCs), white blood cells and other suspended bodies. Previous such coupled simulations were confined to microscale vessels. In order to scale up the simulation up to realistic geometries, higher computational power is required. In this work, authors present multiscale simulation of cardio vascular flows of human by using 4,000 GPUs of Tokyo-Tech TSUBAME2.0 supercomputer. Spatial resolution extends to 5cm down to 10um, and the simulation involves a billion fluid nodes, with 300 million suspended bodies.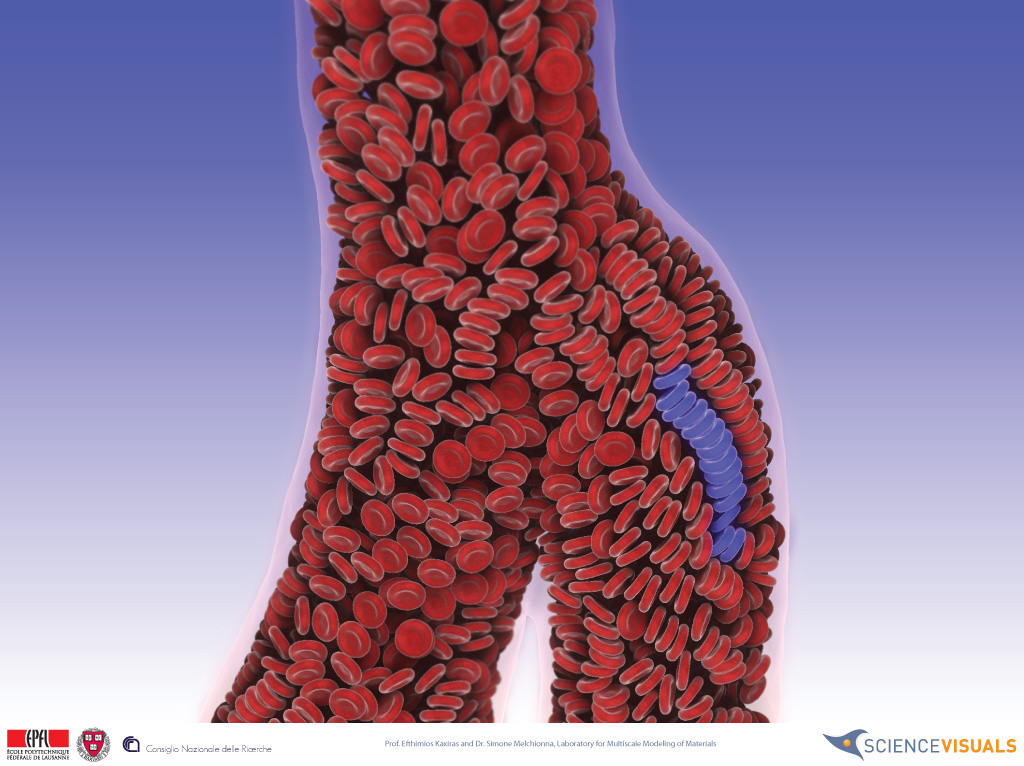
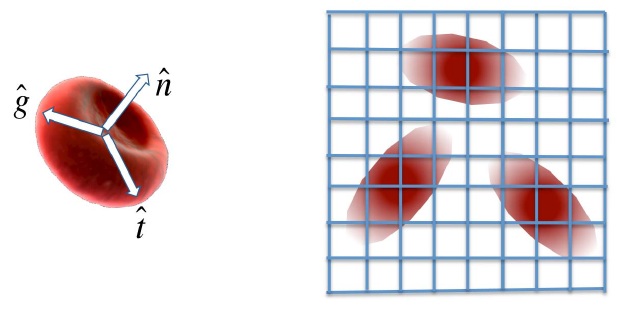
Red blood cells as ellipsoidal particles
Methodology
The multiphysics/multiscale simulation described above is performed with the MUPHY code by authors' group. MUPHY couples Lattice Boltzmann (LB) method for the fluid flow and a specialized version of Molecular Dynamics (MD) for the suspended bodies. A CUDA version of MUPHY has been implemented for the evaluation on thousands of GPUs. For fluid computation, while GPUs are computing elements, CPUs are used for boundary exchange; thus overlapping computation and communication is achieved.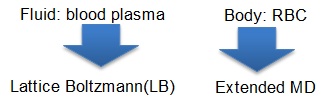
Since our target domain has irregular structure, partitioning the domain among 4,000 GPUs is challenging; for this purpose, a third-party software PT-SCOTCH, the parallel version of SCOTCH graph partitioning tool.
Experimental Results
The performance of the multiscale simulations is measured by using 4,000 NVIDIA Tesla M2050 GPUs on TSUBAME2.0. Since a single node embodies three GPUs, 1,334 nodes are used. The simulations include:
- 1 billion lattice sites for the fluid
- 300 million RBCs
Most computations are performed in single point precision. Used system software packages include CUDA compiler version 3.2, Intel Fortran compiler version 11.1 and OpenMPI version 1.4.2.
As a result, the total performance of about 600TFlops (=0.6PFlops) has been achieved, with a parallel efficiency in excess of 90 percent. Among the computation components, LB components are more than a factor of two better than those authors reported previously. This performance corresponds to simulating a full heartbeat at microsecond resolution in 8 hours.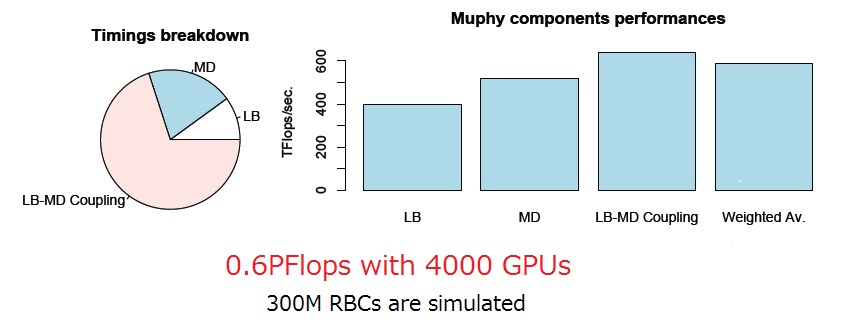
Reference
M. Bernaschi, M. Bisson, T.Endo, M. Fatica, S. Matsuoka, S. Melchionna, S. Succi. Petaflop biofluidics simulations on a two million-core system. In Proceedings of IEEE/ACM Supercomputing '11, Seattle, 2011. (To appear)
![[GSIC] Tokyo Institute of Technology | Global Scientific Information and Computing Center](../../_img/logo_en.jpg)